
Howdy Phage Galaxy Users!
We’ve recently updated our Digestion Toolkit, originally built by our BICH491 student Stephen Crosby, to take advantage of the new plasmid visualization library angularplasmid. This was built into a small workflow which
- handles ligating all of the sequences in a fasta file with a 5′ adapter, a 3′ adapter, and some sequence in between ligands
- optimises only the gene regions for a specific host
- then digests and produces a nice plot of the synthetic plasmid
The workflow is publicly available in the “Shared Data -> Published Workflows” menu.
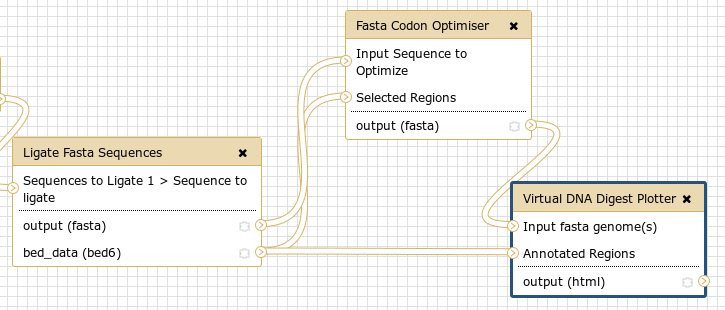
It’s a dead simple workflow which does all of the above steps. I recommend copying it, extending it for your use case (changing the ligation sequences, changing the host for adaptation, etc), and trying it out!
