Restriction enzymes, which cleave DNA at specified sequences, have been critical in the development of biotechnology and genetic engineering. However, the biological role of these enzymes is in restriction-modification (R-M) systems that have evolved to cleave the bacteriophage DNA injected into the cell and thus act as a defense mechanism against phage infection. As a counter-measure, phages have evolved anti-restriction mechanisms that act against the R-M systems. The well-studied bacteriophage P1 of E. coli has two anti-restriction proteins, DarA and DarB (defense against restriction), that act against several type I R-M systems found in E. coli strains. DarA inhibits the endonuclease activity of the restriction enzyme EcoA, whereas DarB prevents inhibits enzymes EcoB and EcoK. Although DarA does not have any homologues and no conserved domains, DarB has recognizable methyltransferase and helicase domains. Moreover, DarB homologues are encoded in mobile DNA elements that encode virulence factors in pathogenic bacteria. Knowledge of the molecular mechanism of Dar function would not only contribute to our ability to use phages as anti-bacterial agents but also illuminate pathways by which disease potential spreads among bacterial species. We aim to use genetic and biochemical analysis to interrogate Dar protein structure and function against against the type I R-M system. The undergraduate student working in this project will be gain experiencw with gene cloning, bacterial physiology, protein purification and biochemical assays. 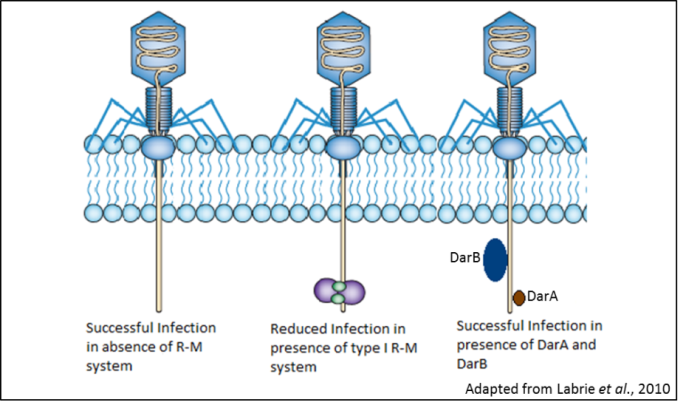
Interested students can contact Dr. Jason Gill for information.
