Another tRNA prediction tool has been added to the CPT Galaxy repertoire: tRNAScan-SE. In version 2.0, the default is set to use the bacterial model for prediction on any input nucleotide fasta. Accompanying tRNAScan-SE is a format conversion script Convert tRNAscan-SE table to gff format, written in-house by Anthony Criscione, that allows this data toLearn More…
Tool Spotlight: Charges
A clean representation of protein primary sequence can do wonders for making a compelling case about protein fold or function. The Charges tool allows you to annotate custom residue combinations with user-specified colors. The output is a scalable vector graphic file that can be used to make high-quality figures from any protein fasta input. PresetLearn More…
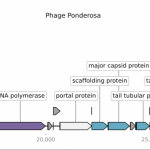
New Tool: Linear Genome Plot
Draw detailed gene maps for your linear phage genome genbank files using Linear Genome Plot. This new tool has flexible options for genome display on a single or multiple lines with coloring feature shapes based on type (gene, regulatory, etc.) or name (such as product). Label source and placement are also set by the user.Learn More…