BICH 464 / GENE 464: Bacteriophage Genomics – 3 credit hours
Bacteriophage Genomics has actually been in operation for over 20 years now. Throughout most of its existence it has been supported by the National Science Foundation. The general idea is to offer TAMU life science majors an opportunity to learn about modern genomics by real hands-on research experience in which they annotate a new phage genome. What does “annotate” mean? Well, when you first get your genome, it will be just a long string of the four DNA bases (A, G, C, and T), probably 50,000 to 250,000 bases in length.
When you are finished at the end of the semester, you will have identified all of the phage’s genes that code for proteins and functional RNA’s, and many accessory features as well. These features will be annotated on the phage DNA sequence, which makes the genome record useful to other researchers.
This annotated genome record will be deposited into GenBank at NCBI and serve as a central element in a final paper that will be submitted in lieu of a final exam. These phage genomes will also serve as the basis for future peer-reviewed publications, and students will be listed as co-authors papers that contain their genomes.
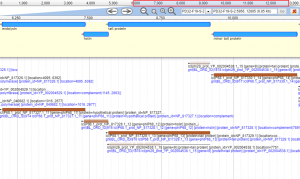
The central concept for BICH 464 is that if a student gets “ownership” of a novel genome, instead of a cookbook exercise, that sense of ownership will be a source of motivation and drive that will greatly enhance the learning process (as well as generate a scientific work-product!). Bacteriophages, which are simply the viruses of bacteria, were chosen as the organisms of choice for the simple reason that they have relatively small genomes, typically on the order of 100,000 base pairs of DNA or ~100 genes. That may sound like a lot, but it is about two orders of magnitude less than a typical bacterial genome, and 10,000-fold less than a typical eukaryotic genome, both of which would be just too large for a student to handle, especially in one semester. Also, we know each phage genome has to have genes for things like the viral capsid, tail, DNA packaging and host lysis, which makes it a bit easier to assign gene function. In any case, the system has worked well for a long time.
Course Content
So what actually happens in the course, and what would you be expected to do? The course has three major phases: lecture, lecture/computer lab, and finalizing the genome. During the first phase, the classroom sessions are dedicated to an intensive survey of basic bacteriophage biology. You will learn what phages are and how they work at the molecular level. The second phase starts when you get assigned your new phage genome. At that point, time is divided between lectures on phage biology and computer sessions devoted to genome annotation and learning bioinformatics methods and tools. In the third phase, sessions are increasingly dedicated to the computer instruction and workshops focused on actual issues arising during the annotation of your particular genome, culminating in the completion of your annotation project and your final paper.
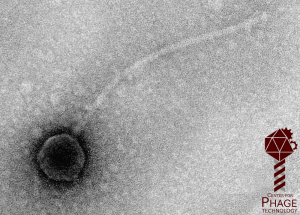
In the early years of the course, we used to do the sequencing of the new phages ourselves, which occupied most of the course. However, in modern biology, whole genome sequencing is done commercially with “next generation” technologies, and the real scientific bottleneck is the annotation. That’s where you come in…
Interested?
You are officially encouraged to drop us a line. Course size is typically limited to 20 students, and it is best if you have had some preparation for the microbiology and molecular biology concepts that are a central part of the course material. Having taken Genetics (GENE 302) is strongly recommended, and some combination of microbiology (e.g., BIOL 351), molecular genetics (e.g., BICH/GENE 431), biochemistry (e.g., BICH 410/411/440/441) or bacterial genetics (BIOL 406) will also help provide a good background. However, we can be flexible, depending on your background and academic goals. Starting in 2021, the course has been cross-listed to the Genetics program, so it can now be taken as either BICH 464 or GENE 464. We also accept graduate students in the course, and most programs will allow grad students to take a limited number of 400-level courses as part of their degree plans. The simplest approach is to email one of the course staff:
| Position | Name | Contact |
| Instructor | Jason Gill | jason.gill@tamu.edu |
| Secretary | Mrs. Daisy Wilbert | daisy@tamu.edu |
To apply, you will need to submit an unofficial transcript (.pdf) to Daisy at daisy@tamu.edu. You can also come by room 308 in Bio/Bio and inquire directly to Daisy. In either case, you will get a quick response!
