Salmonella bacteria are often health hazards, associated with a million foodborne illnesses per year in the US. Phages have been used to identify Salmonella species and may also be useful in therapy and prophylaxis of Salmonella infections. The phage FelixO1 was first used in 1943 by Felix and Callow as part of a “phage-typing” system for the identification Salmonella typhi. 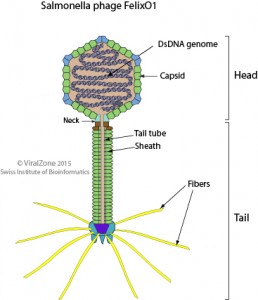
FelixO1 is unusual among Salmonella phages because of its specificity, attacking >98% of Salmonella strains but less than 2% of other Enterobacteriaceae. Its genome was sequenced and annotated, revealing it to be a distant relative of the classic E. coli myophage T4. Despite close inspection and analysis, however, we have not been able to identify its lysis control genes (holin and potentially anti-holin). Because FelixO1 is considered to be one of the three most “famous” Salmonella phages, the others being the classic temperate podophage P22 and the “lysogenic conversion” phage epsilon15, it is important to discern whether or not it has a new paradigm for lysis control. In this project, we would start by constructing a mutant library and screening for lysis defects. It would involve learning how to culture bacterial strains, plate bacteriophages, conduct mutagenesis protocols, screen for nonsense mutants, sequence DNA, assess lysis phenotypes, and do complementation tests. This is an ideal starter project for someone who has the interest in getting into bacteriophage research. If the progress allows, the project may be expanded into a generalized hunt for all essential genes and also for finding host genes required for FelixO1 growth.
Contact Prof. Ry Young if you are interested.
