Spanins are bacteriophage lysis proteins responsible for disruption of outer membrane (OM), the final step of Gram-negative host lysis. The absence of spanins results in a terminal phenotype of fragile spherical cells. The phage T1 employs a unimolecular spanin (US) gp11 that has an N-terminal lipoylation signal and a C-terminal transmembrane domain (TMD). Upon maturation and localization, gp11 ends up as an OM lipoprotein with a C-terminal TMD embedded in the inner membrane (IM), thus connecting both the membranes as a covalent polypeptide chain. Unlike the two component spanins (2CS) encoded by most of other phages including lambda, the u-spanins haven’t been studied extensively yet. The aim of my research is to investigate the molecular mechanism behind US function and further gain insights into phage lysis as well as the bacterial cellular envelope.
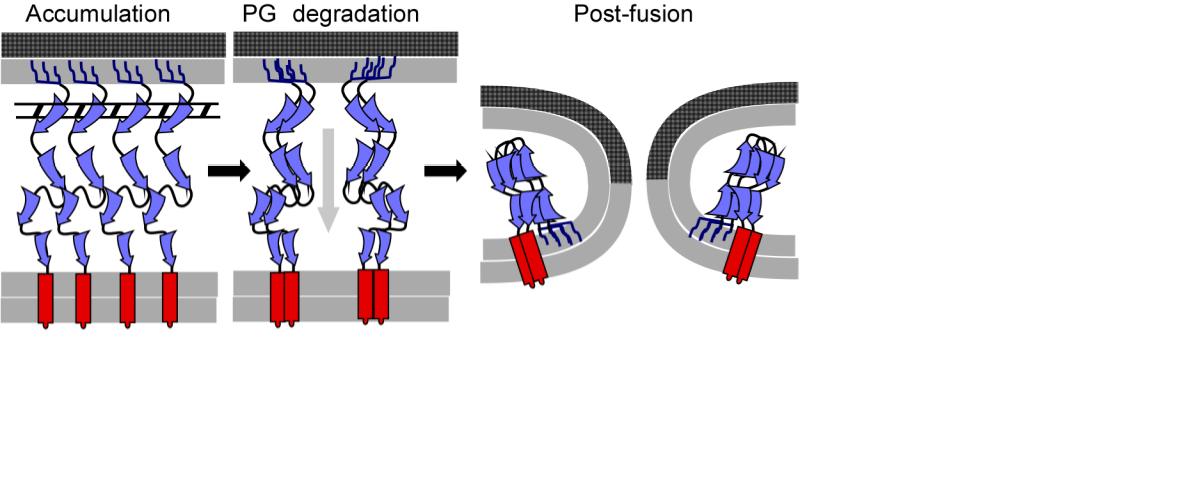
We used a detailed bioinformatics based, biochemical and genetic approach to study gp11. Using the ability of gp11 to complement the lRzRz1 lysis defect, we showed that both the OM and IM localization signals were true and necessary for gp11 function. Furthermore, 14 lysis-defective single missense mutants, distributed throughout the periplasmic domain of gp11, were isolated from a mutant library created with error-prone PCR mutagenesis. Fluorescence spectroscopy time lapse videos using gp11-gfp showed gp11 accumulating in distinct punctate foci, suggesting localized oligomerization within the peptidoglycan meshwork. Our current model for gp11 function hypothesizes that a conformational change occurs in spanin complexes after peptidoglycan degradation leading to fusion of the IM and OM.