Genome annotations are constantly improving. In the process of working on an annotation, or re-annotation, project, you can accumulate various Genbank file versions for the same organism. When you need a wholesale comparison of the number, position/length, and names or notes for each CDS feature, try using our new Compare Genbanks tool.
The tool will take two Genbank files for the same organism and compare the CDS features pairwise, logging the similarity, start/end position, names, and notes for each. Those results are output in two files. The first gives an overall picture of the comparison. Name changes from ‘hypothetical’ to anything else are also counted, helping you get an idea of the improvement in the annotation.
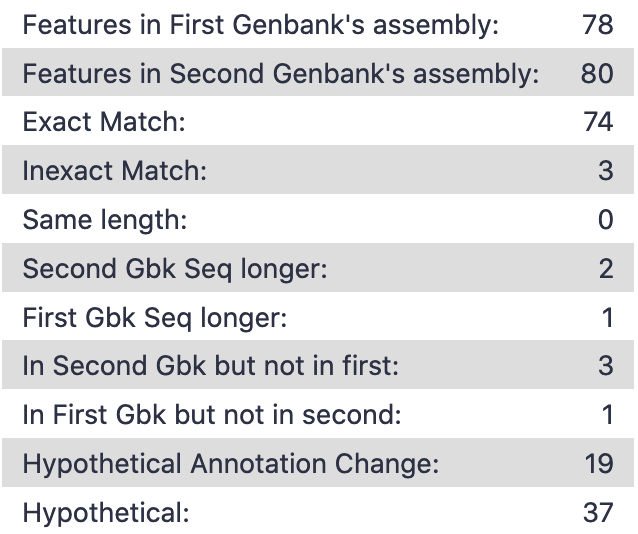
The second output does a more fine-grained presentation of the features for the entire genome. In this format, you can see exactly where the changes have occurred, and see where to double check your work as needed.
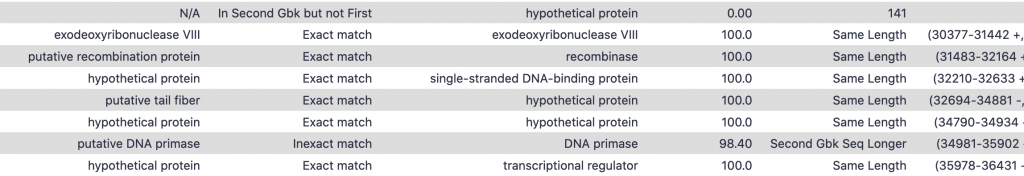
Whether you are checking an automatic annotation against a manual annotation, comparing a new assembly with an old one, or simply lost track of which file is the latest, the Compare Genbanks tool will help you re-organize and focus on the most important places in your assembly.
Written and wrapped for Galaxy by Curtis Ross and Anthony Criscione, the Compare Genbanks tool can be found under CPT: Genomic Overview. Please email the support team if you have any questions!
