The final step in a bacteriophage infection cycle, lysis and release of progeny virions from the host, requires destruction of the bacterial cell envelope, which in Gram-negative bacteria, includes the cytoplasmic membrane, the cell wall (peptidoglycan), and the outer membrane. For the simple, ubiquitous lytic single-strand RNA and DNA phages, lysis is effected by the expression of a single gene. There are three general classes of these phages: the ssDNA microviruses and two classes of ssRNA phages, the leviviruses and the alloleviruses. By studying how these lysis genes make the host undergo lysis, we expect to advance our understanding of how bacteria synthesize and maintain their cell envelope and thus potentially open the way for developing new antibiotic strategies.
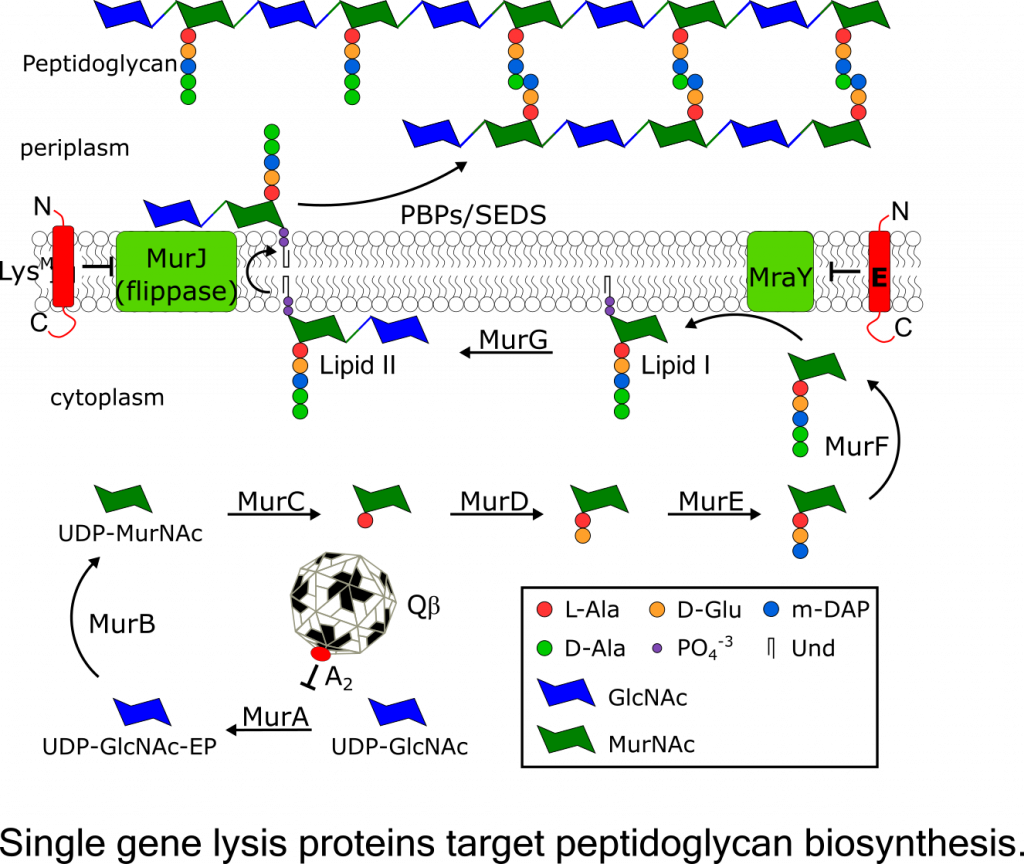
Previously, three different single gene lysis systems identified in the well-studied paradigm phages representing the three phage classes: E of the microvirus fX174, A2 of the allolevivirus Qb, and L of the levivirus MS2. Work in done in the Young lab has shown that A2 and E function as specific inhibitors of enzymes in the pathway for murein precursor biosynthesis, MurA and MraY, respectively. The third prototypic lysis protein L causes lysis by an unknown mechanism and is currently being investigated by my team.
Recently, we discovered a fourth mode of single gene lysis system, the lysis protein LysM from the Escherichia coli phage M. LysM was also shown to target peptidoglycan biosynthesis by inhibiting MurJ, the essential lipid II flippase. This recent discovery raises the possibility that ssRNA phages have evolved proteins that target other essential steps in peptidoglycan biosynthesis and maintenance. Thus, understanding the lysis mechanisms of ssRNA phages might facilitate the discovery of novel targets for antibiotic development.