A new version of the Structural annotation workflow has been released! The v2021.02 workflow has an updated ShineFind tool, which searches upstream of the start codon of an open reading frame for Shine-Dalgarno sequences.
* Structural Workflow: PAP Structural Workflow v2021.02
Notable updates: latest version of the ShineFind tool improves selection of spacing between the start codon and the ‘best’ Shine-Dalgarno sequence
Shared by: cpt-workflows
Whereas a previous version of ShineFind used an inclusive nucleotide window within which the Shine-Dalgarno had to be located, the latest update now allows users to specify the allowed gap between the start codon and the end of the Shine-Dalgarno sequence. In the structural workflow v2021.02, that gap is preset to 3-17 nts, and it can be adjusted at runtime if needed. The standalone tool default also uses the 3-17 nts.
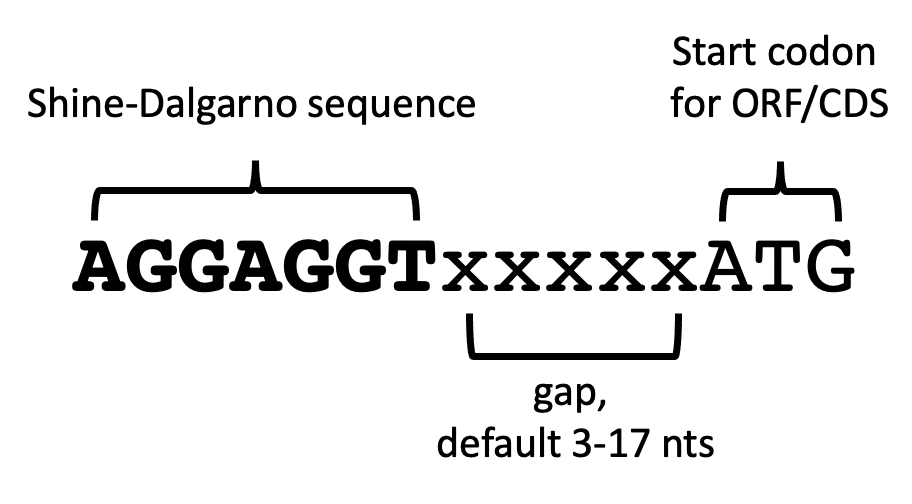
What if there is more than one possible Shine-Dalgarno sequence that has an allowed gap distance? First of all, ShineFind can output all the possible matches within the set parameters. However, we usually use the ‘Only report best hits’ option, which will output a single Shine-Dalgarno per input coding sequence. Previous versions of ShineFind chose the *first* (or furthest upstream) sequence that matched the requirements. Now, ShineFind selects the one with the smallest gap.

The latest version of all published workflows can be used or imported at Shared Data>Workflows. Have questions? Email the support team!
