Past Project Leaders and Student Researchers
Project LeaderJolene Ramsey, Ph.D.
Phage exit is a tightly regulated process. In gram-negative bacterial hosts, phage must cross three cell barriers, the inner and outer membrane and the peptidoglycan cell wall, to release phage progeny. To understand the proteins that govern those processes, our team studies lysis proteins in both well-studied canonical phage systems (lambda, Mu) and less known phages as well. Here we apply molecular, biochemical, and genetic approaches to tease out the pathways leading to lysis. As the Center for Phage Technology mandate states, we are interested in finding and characterizing novel phage from the environment that target relevant bacterial hosts. Our host panel is continuously being updated based on the needs in the community; these are not your garden variety E. coli. These projects could be the seed for industry partnerships and often extend from or flow into the BICH 464 Bacteriophage Genomics course. |
491 Students under Jolene Ramsey (2018 – 2022)

Holt, Ashley |
Martin, Cody |
Awah, Adey |
Tomaszewski, Adam |

Snowden, Jason |
Nittolo, Toni |
Corban, James |
Torres, Ruben |

Sullivan, Teresa |
Panin, Nick |
Mota, Andrea |
Reichert, Dacia |
Publications
- Holt, A, Cahill, J, Ramsey, J, Martin, C, O’Leary, C, Moreland, R, Maddox, L et al.. Phage-encoded cationic antimicrobial peptide required for lysis. J Bacteriol. 2021 Aug 2;JB0021421. doi: 10.1128/JB.00214-21. PubMed PMID:34339297.
- Corban, J and J Ramsey. Characterization and complete genome sequence of Privateer, a highly prolate Proteus mirabilis podophage. PeerJ. 2021 Feb 10;9:e10645. doi: 10.7717/peerj.10645. PubMed PMID:33614267 PubMed Central PMC7881722.
- Clark, J, Awah, A, Moreland, R et al.. Complete Genome Sequence of Vibrio natriegens Phage Phriendly. Microbiol Resour Announc 2019 Oct 3;8(40):e01096-19. doi: 10.1128/MRA.01096-19. PubMed PMID:31582448 PubMed Central PMC6776788.
- Cahill, J, Rajaure, M, Holt, A, Moreland, R, O’Leary, C, Kulkarni, A et al.. Suppressor Analysis of the Fusogenic Lambda Spanins. J. Virol. 2017;91 (14):e00413-17. doi: 10.1128/JVI.00413-17. PubMed PMID:28468876 PubMed Central PMC5487574.
- Cahill, J, Rajaure, M, O’Leary, C, Sloan, J, Marrufo, A, Holt, A et al.. Genetic Analysis of the Lambda Spanins Rz and Rz1: Identification of Functional Domains. G3 (Bethesda). 2017;7 (2):741-753. doi: 10.1534/g3.116.037192. PubMed PMID:28040784PubMed Central PMC5295617.
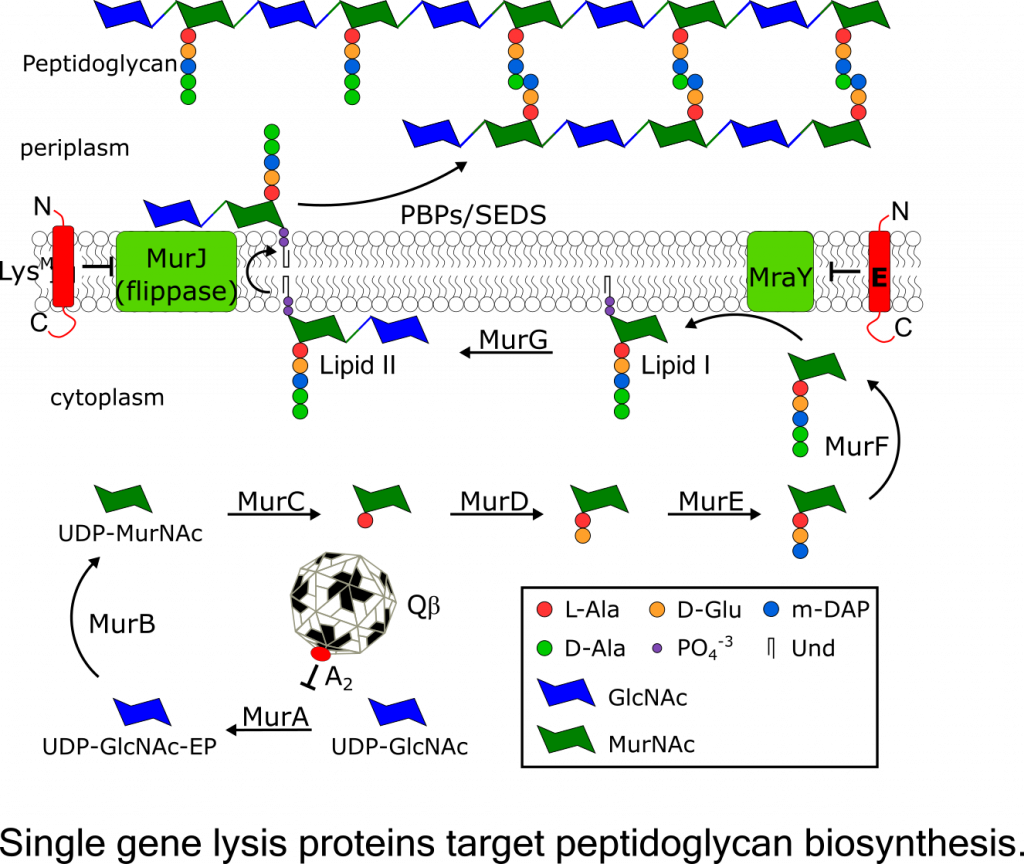
Project LeaderKarthik Chamakura, Ph.D. Single Gene Lysis Systems The final step in a bacteriophage infection cycle, lysis and release of progeny virions from the host, requires destruction of the bacterial cell envelope, which in Gram-negative bacteria, includes the cytoplasmic membrane, the cell wall (peptidoglycan), and the outer membrane. For the simple, ubiquitous lytic single-strand RNA and DNA phages, lysis is effected by the expression of a single gene. There are three general classes of these phages: the ssDNA microviruses and two classes of ssRNA phages, the leviviruses and the alloleviruses. By studying how these lysis genes make the host undergo lysis, we expect to advance our understanding of how bacteria synthesize and maintain their cell envelope and thus potentially open the way for developing new antibiotic strategies. Previously, three different single gene lysis systems identified in the well-studied paradigm phages representing the three phage classes: E of the microvirus fX174, A2 of the allolevivirus Qb, and L of the levivirus MS2. Work in done in the Young lab has shown that A2 and E function as specific inhibitors of enzymes in the pathway for murein precursor biosynthesis, MurA and MraY, respectively. The third prototypic lysis protein L causes lysis by an unknown mechanism and is currently being investigated by my team. Recently, we discovered a fourth mode of single gene lysis system, the lysis protein LysM from the Escherichia coli phage M. LysM was also shown to target peptidoglycan biosynthesis by inhibiting MurJ, the essential lipid II flippase. This recent discovery raises the possibility that ssRNA phages have evolved proteins that target other essential steps in peptidoglycan biosynthesis and maintenance. Thus, understanding the lysis mechanisms of ssRNA phages might facilitate the discovery of novel targets for antibiotic development. |
||||||||
491 Students under Karthik Chamakura (2016- 2021)
Publications
|
Former Student Researchers
| 2010 | |
| Johnson, Bryan | graduate school |
| Shrestha, Ritu | PhD program in Biology, Texas A&M University |
| 2011 | |
| Kaspar, Justin | PhD program in Interdisciplinary Biomedical Sciences at University of Florida |
| Lessor, Lauren | Research Assistant, CPT program, Texas A&M University |
| Mohan, Sheba | Physician Assistant, DFW |
| Tsau, Joshua | MD program, UT Houston Medical School |
| 2012 | |
| Dachowski, Michael | MD program, University of Texas Medical School, Houston |
| Herrera, Oscar | Clinical Pathology Laboratories, San Marcos |
| Lawler, Jessica | PhD, Dept. of Biological Chemistry & Molecular Pharmacology, Harvard Medical School |
| Migl, David | PhD program in Biophysics, Harvard University |
| 2013 | |
| Brahmbhatt, Kirtan | MD program, Texas A&M University Health Science Center |
| Kulkarni, Aneesha | PhD program in Molecular Biology focusing in immunology, San Jose State University |
| Park, Katherine | MD program, Baylor College of Medicine |
| Rasche, Helena | System Administrator for UseGalaxy.eu |
| Simpson, Jacob | MS Health Administration TAMU HSC |
| 2014 | |
| Edwards, Garrett | PhD program in Biochemistry, University of Colorado, Boulder |
| Khatemi, Brontee | the director of education at Sylvian Learning Center, Dallas, TX |
| Ladzekpo, Tsonyake | Medical Scribe at PhysAssist Scribes, Inc., Houston, TX |
| Luna, Adrian | PhD program in Biomedical Sciences, University of New Mexico |
| Snowden, Jeffrey | PhD program in Micro-logy & Cell Biology, University of Texas Medical Branch, Galveston |
| 2015 | |
| Church, Kaira | certification in Clinic Lab, UT Health Science Center in San Antonio, TX |
| Lancaster, Jacob | General Surgery Resident, University of Kansas Medical Center |
| Bernal, Christopher | Masters program in BIMS, Texas A&M |
| LeSage, Kayla | Masters program in BIMS, Texas A&M |
| Mijalis, Eleni | MD program at Louisiana State University – Shreveport |
| Crosby, Stephen | PhD program |
| Maughmer, Cory | Programming Analyst, Lone Star Analysis |
| 2017 | |
| Min, Lorna | MD program Baylor College of Medicine |
| 2018 | |
| Tran, Jennifer | PhD program, University of Wisconsin Madison |
| Moreland, Trey | MD program, University of Texas Rio Grande Valley |
| O’Leary, Chandler | MD program, University of North Texas – Fort Worth |
| Torres, Ruben | PhD program, Vanderbilt University |
| 2019 | |
| Holt, Ashley | MD/PhD program, University of Wisconsin-Madison |
| Nittolo, Toni | Lab Technician, Texas A&M University |
| 2020 | |
| Corban, James | PhD program, University of Wisconsin-Madison |
| Snowden, Jason | PhD program, Texas A&M University |
| 2021 | |
| Martin, Cody | PhD program, University of Wisconsin-Madison |
| Tomaszewski, Adam | Research Assistant, Lynntech Inc. |
| Mota, Andrea | Post-baccalaureate at the National Institute of Health (NIH) |
| 2022 | |
| Sullivan, Teresa | |
| Reichert, Dacia | Texas A&M University |