8. CPT GO Annotations¶
8.1. CPT GO¶
The CPT has built a system for making gene ontology (GO) annotations. This system is highly integrated with Apollo, you will see the results of your annotations within the Apollo interface. GO Terms are used to make GO Annotations.
Before making a GO annotation, you should know:
- In general, the term you will use to describe the feature.
- The specific evidence you have for your annotation (e.g. Blast hit ID.)
8.2. Making a GO Annotation¶
Right click on the feature you wish to annotate in the User Created Annotation track.
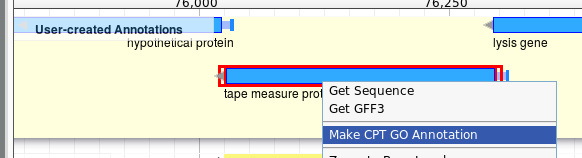
This will open a window with which to create annotations. On the left is the entry form. On the right we see a list of GO annotations that have previously been made to this feature.
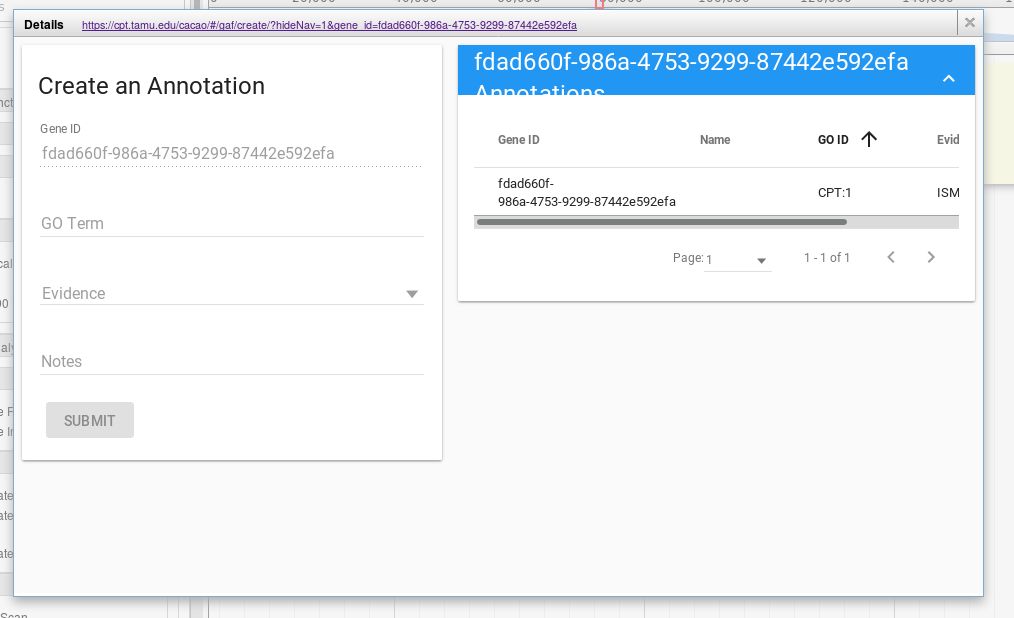
This interface will permit you to make GO annotations that are annotated with evidence codes. Keeping track of this chain of evidence is key to good science.
The first field, gene ID, is filled out for you. The second field, your first decision, is the GO term you wish to use. You can type freely into this box, our system will attempt to find a matching, pre-existing GO term. If you cannot find a term that describes your gene well, you can enter free text into the field.
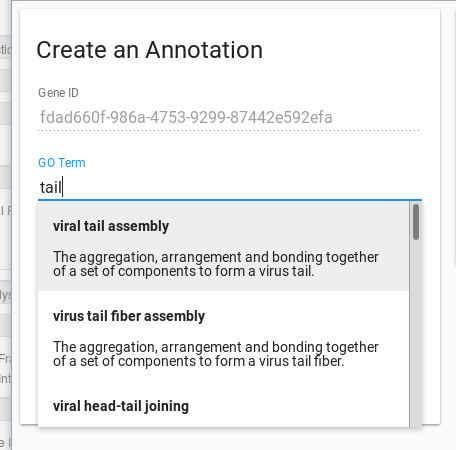
Once you have decided on your term, next you need to specify the evidence you are using. There are three choices for acceptable evidence:
BLAST: This form of evidence requires annotating the database and the database identifier. If your BLAST hit is from the canonical database, its description will look like
lcl|NC_000866.4_prot_NP_049708.1_94 [gene=nrdC.10] ...; for the database you will choose Canonical, and the identifier will be NP_049708.1 (the second accession number, which is the protein accession). Non-canonical BLAST hits will have a description likeUniRef90_E3T7X9 Uncharacterized protein n=7 Tax=Viruses TaxID=10239.... In this case the database is UniRef90, and the idnetifier is E3T7X9.TMHMM: Here you have a TMHMM hit to your protein, and from this hit you can infer some aspect of functionality based on this evidence.
Genomic Context: This evidence type relies on the presence of genes with specific functionality nearby. A good example of this case are lysis genes. The presence of an i-spanin that you can identify often implies an o-spanin just downstream. You may only have some functional domain evidence from InterPro for your gene and the presence of the upstream I-Spanin with which to make an annotation. You can formalize this as evidence due to the genomic context of the upstream feature.
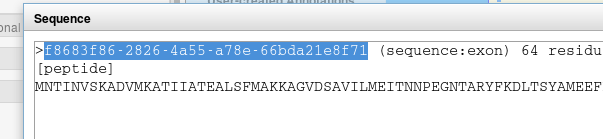
When annotating, you will need to get a feature ID. The easiest way to do this is to click “Get Sequence”, and then to copy the long ID that appears in the fasta header.
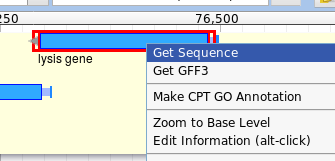
Make some brief notes about how you came to your conclusion, similar to how you use the notes field in Apollo. When you’re done, click “Submit” and close the popup.
