5. Structural Annotation Workflow [BICH464 Student Version]¶
5.1. Prerequisites¶
- Download your fasta file from the Google Drive folder 2018 Student Genomes.
- Upload your fasta formatted file to Galaxy. - The instructions for uploading data to Galaxy can be found in the Introduction to Galaxy tutorial.
5.2. Workflow¶
This workflow will run your genome through two automated gene callers, MetaGeneAnnotator and Glimmer3. A completely naive set of ORFs will also be generated by a tool called Sixpack, in case you believe you find a gene not found by the gene callers. Lastly, tRNA and terminator finding tools will be run.
Open the workflow in Galaxy.
Select import at the top right, in green.

Fig. 5.1 NB: Import workflow “PAP 2017 Structural (v8.9)” You need to use the green “+” button at the right to import the workflow
Once you’ve imported it, you’ll want to start using this workflow
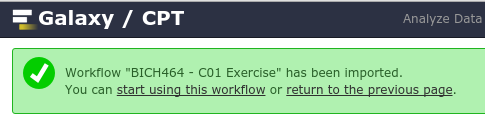
Again, using the dropdown to actually Run workflow
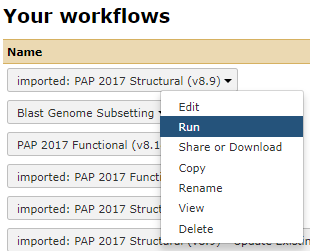
Fig. 5.2 NB: This is a generic image, your workflow may have a different name.
Specify the following:
| Step | Field | Data |
|---|---|---|
| Step 1: Input Dataset | Fasta Genome | Your phage’s Fasta Genome |
| Step 25: Create or Update Organism | Genus / Species | Genus and Species, if available. |
Execute
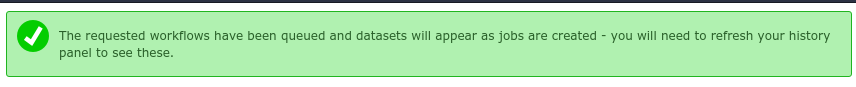
If everything went successfully, you’ll see a notice, you should follow the instructions
5.3. Gene Calling¶
This workflow has now loaded your genome and the tracks of gene call evidence into Apollo. From there you will need to go through the genome and select the likely gene calls. Remember that the three gene callers are correct maybe 90-95% of the time. Your task is to examine each likely gene region, and select the best call.
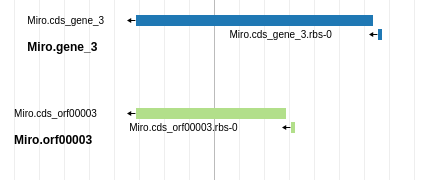
Fig. 5.3 Blue is probably a better choice than green in this case, because it has higher genomic coverage and a Shine Dalgarno site identified.
You will need to “Create » Gene” for every likely gene in your genome. It is better to over-call your genome (call more genes than are likely to be there), than to under-call genes and have to go back and re-run blast and related analyses. You will need to complete this process before you can start on the functional annotation.
