Exercise 3: NCBI Blast¶
Notes¶
- Please follow steps closely
- Raise your hand if you have questions
- This is available online if you want click-able links at
https://cpt.tamu.edu/bich464/C1-exercise.html
Exercise¶
Here is your protein, you’re tasked with identifying it!
>unknown_protein
VVKSSGVRQPFDKEKIYKVLKWACDGHNIDVRAFLENVLELIRDGMTTKQIQRIAAIKYA
ADHISVKEPDWQYVASNLEMFALRKDVYGQFDPIPFYDHIVKMVEAGKYDKEILEKYSKQ
DIQVFERAIDHDKDFEFSYAGSQQLIGKYLVQDRDTGEIFETPQYAFMLIAMCLHQEETG
AQVTHIVDFYNAISDRKLSLPTPIMAGVRTPTRQFSSCVVIESGDSLGSLNAVTSAIKVY
ISQRAGIGVNAGHIRAMGSKIRGGEAVHTGVIPFWKIQTAVKSCSQGGVRGGAATLYYPF
WHLEVENLLVLKNNKGVEENRVRHLDYGVQLNQLMYKRLMNRDYITLFSPDVANDRLYDL
BLAST Steps¶
The NCBI BLAST website offers several types of BLAST queries.

Blast queries available
We’ll be doing a Basic BLAST with Protein Blast. You’ll need to
Paste in your Query Sequence, the unknown protein. Then choose
nr for Choose a Search Set
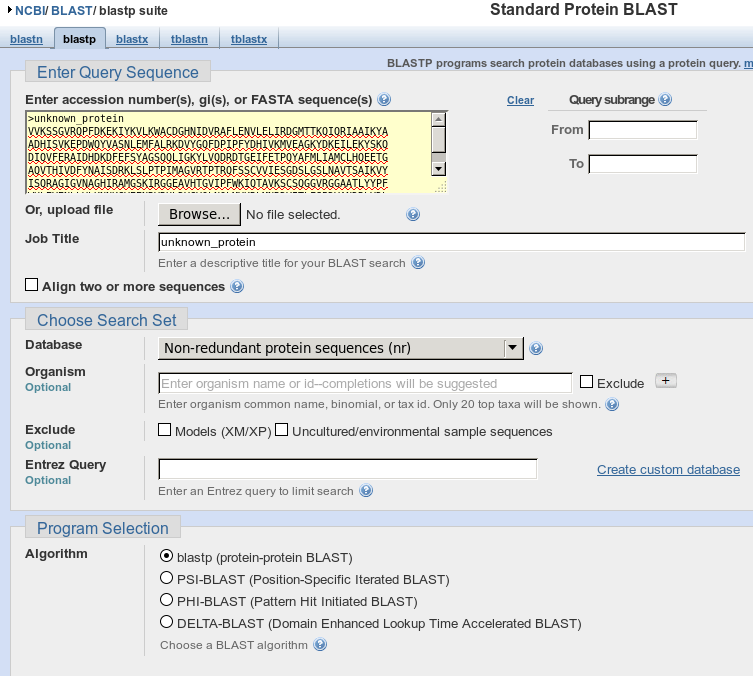
You’re ready to blast! Hit the button and be patient

Run¶
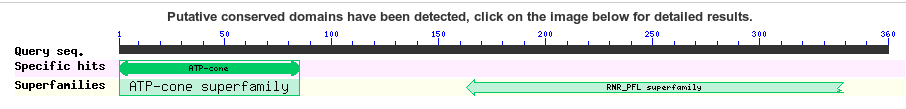
You will see this when blast starts up. Blast has identified a domain in your protein. This can be informative to your annotation and naming process.
If you click on it you can read more about the domain.
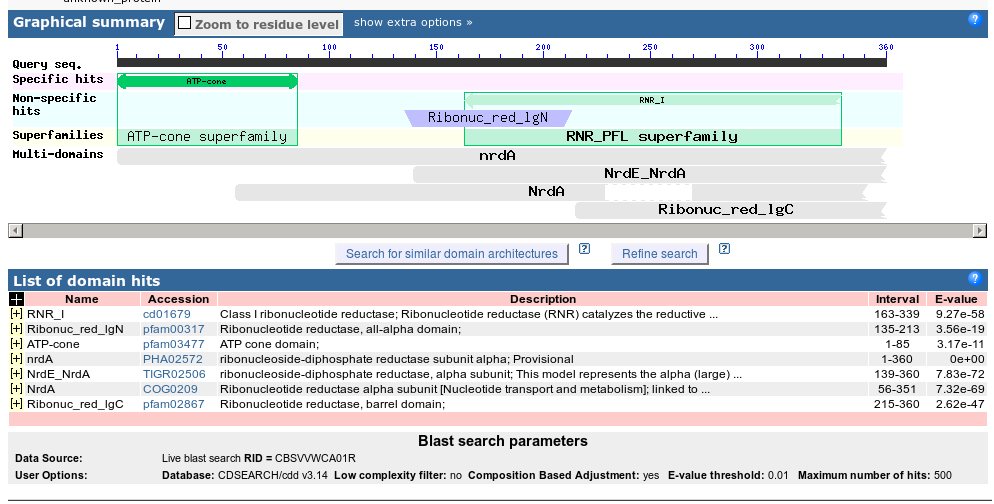
The Conserved Domain Database (CDD) website contains information on different protein domains.
Finished Blast¶
There are several sections to the BLAST output on the web:
- Graphical summary of the matches
- Hit list
- Alignments
Graphical Summary¶
This simply shows you where regions from other proteins hit aligned with your query protein.
Nice hit table covering our blast results
Hit List¶
Here we can see an overview of the database hits. The table is sorted by E-Values, the expectation values.
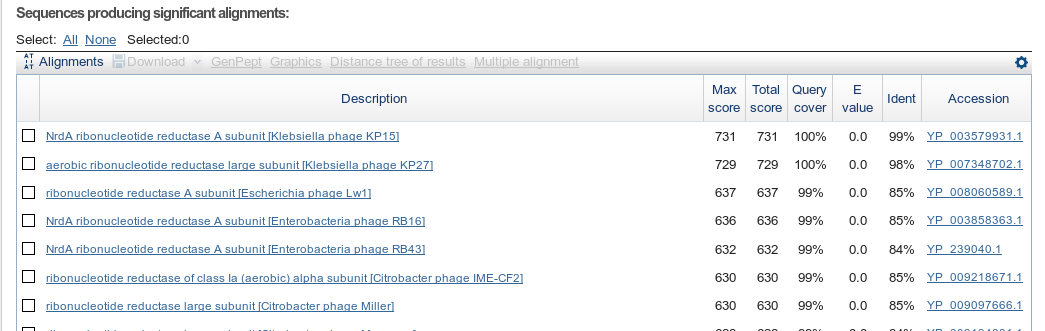
List of individual hits
When looking at blast results on the web, your main goal is usually to figure out the identity of a protein. Here we see lots of blast hits to NrdA or ribonucleotide reductases. Why are they so sure? Since NCBI doesn’t expose any further levels of evidence, we dig through the blast results and see...
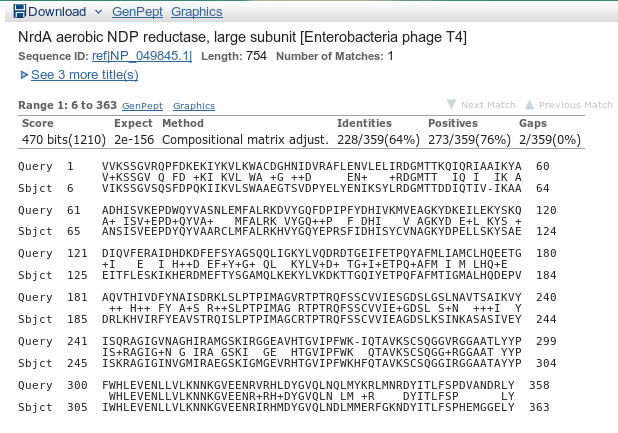
A hit to T4’s NrdA! That’s an extremely good indicator of the identity of your protein (again, in absence of real, wet-lab experiments.)
Completing the Exercise¶
- At the top of the page is the “Download” button
- Select Hit Table(csv) to download a copy of all the hits
- Upload this to Galaxy
- Run 464 C02 - evaluate
How to Use This Information¶
We will be doing blast from within Galaxy and viewing the results. The workflow will be somewhat different from what you’ve learned how to do in this exercise, however the underlying theory is the same. You have a query protein and you’re searching through all the other proteins in the world for similar results.
