Class 3 Slides: Annotation¶
Protein Annotations¶
Triaging Your Phage Proteins¶
- Hypothetical Novel Proteins
- No BLAST hits
- No domain hits
- Hypothetical Conserved Proteins
- BLAST hits to proteins without known function
- No domain hits
- Annotated Protein
- BLAST hits to proteins with annotated function
- Domain hits
Filtering Results¶
- BLASTP results
- Which database?
- Does it make sense?
- Domain results
- How specific/meaningful?
- Does it make sense?
Making a Protein Annotation¶
- An annotation is your prediction of protein role/activity
- Annotations should be useful
- A useful annotation is accurate and specific
- Usually better to trade specificity for accuracy
- More general annotation if not clear about specific function
- Rationale for annotation in the Notes field.
- Make notes!
Apollo¶
Workflow C Output¶
- Turn on the following tracks:
- InterProScan
- BlastP agaisnt Canonical Phage DB
- BlastP against UniRef90
BLAST Against Canonical Phage DB¶
- Only well-understood “canonical” phages are included
- Hits to most/all genes can indicate highly related phages
- Few/no hits in this track implies your phage is not related to a well studied phage
BLAST Against Canonical Phage DB¶

Hits to canonical phage DB
BLAST Hit Information¶
- Click on a blast hit
- Score field contains the e-value
- lower is better
- reflected in track colour
- Description may/may not be informative. Depends on quality of annotation in database.
- May need to go to NCBI and find genome for more information on a gene
- If you do this often, contact Eric and this can be automated.
BLAST Against UniRef90 DB¶
- UniProt is a curated protein database
- Uses evidence-based annotations (NCBI/NR has no such restrictions)
- UniRef90 is database of UniProt proteins clustered with highly related proteins (>90% similarity over >50% of length)
- Zoom in + hover over hits to see names/information
BLAST Against UniRef90 DB¶
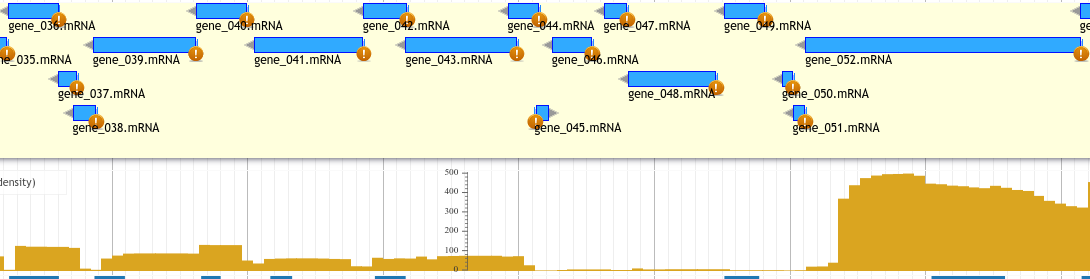
Hits to UniRef90
Analysing UniRef90 Hits¶
- Zoom in to see hits
- Even if no canonical phage hits, should have uniref hits
UniRef90 Hits¶
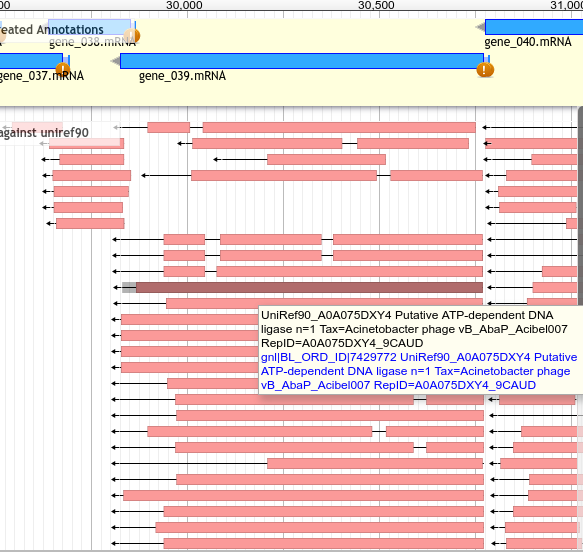
Hits to UniRef90
InterProScan¶
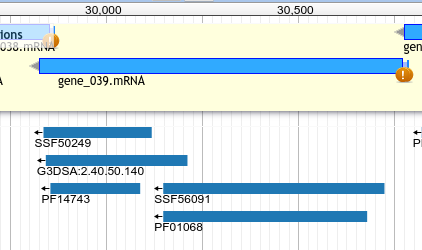
Hits to InterProScan
- Searches for conserved domains in member databases
- Predict protein function based on domains. Beware domain swapping!
InterProScan Hits¶
- If the domain is part of InterPro, will have an
InterPro:IPR#####Dbxref entry. - Searching the Name or Dbxref fields in google will yield results
Annotating a Gene 2¶
- Do NOT fill out mRNA
- Under Attributes, specify a product and a product name
What Should I Annotate Protein X?¶
- No hard conventions for what exactly to name protein
- Look at BLAST results, common to see six homologs of the same protein with 4 different names
- All are technically correct

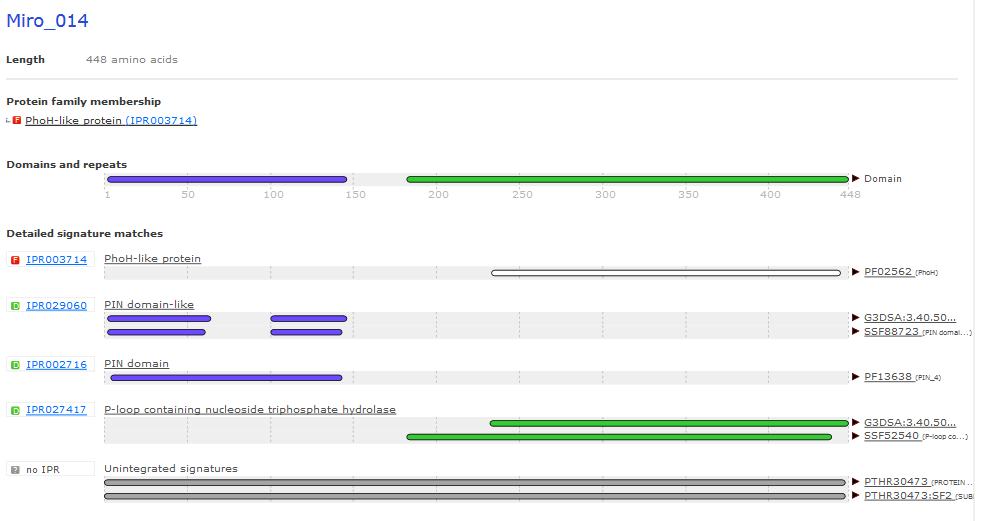
A Tricky Annotation: Miro gp14¶
- 450 AA protein
- Has BLAST hit to PhoH-like protein
- InterPro shows PhoH domain and a PIN domain
- PhoH is an ATPase involved in phosphate starvation response
- PIN domains are ribonucleases, sometimes part of TA system
- Is this really PhoH?
Miro_014 InterProScan Results¶
The E. coli PhoH protein record¶
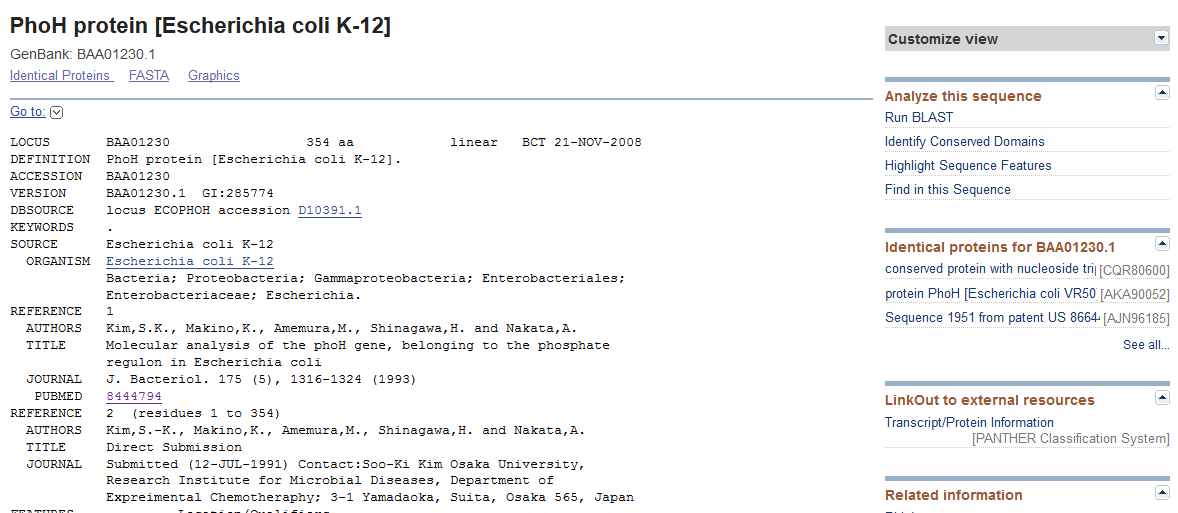
Grab the FASTA protein sequence by clicking “FASTA”, then run InterProScan and BLAST. Note that the “real” PhoH is 100 aa shorter than gp14.
BLAST on Real PhoH¶
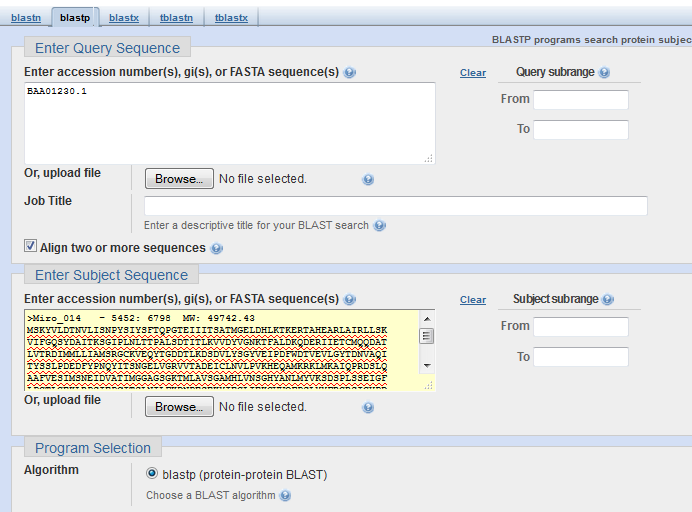
Here we’re using NCBI’s BLAST to do a pairwise alignment.
BLAST Results from PhoH¶
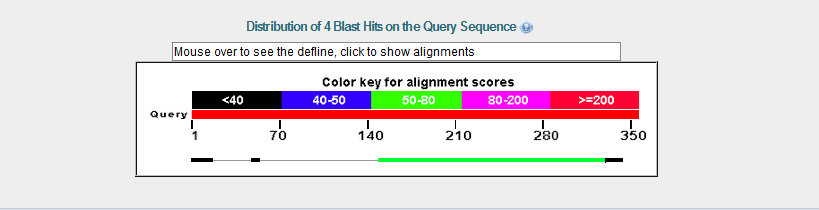
The BLAST alignment shows that a large domain is different in the real PhoH.
InterPro Results from PhoH¶
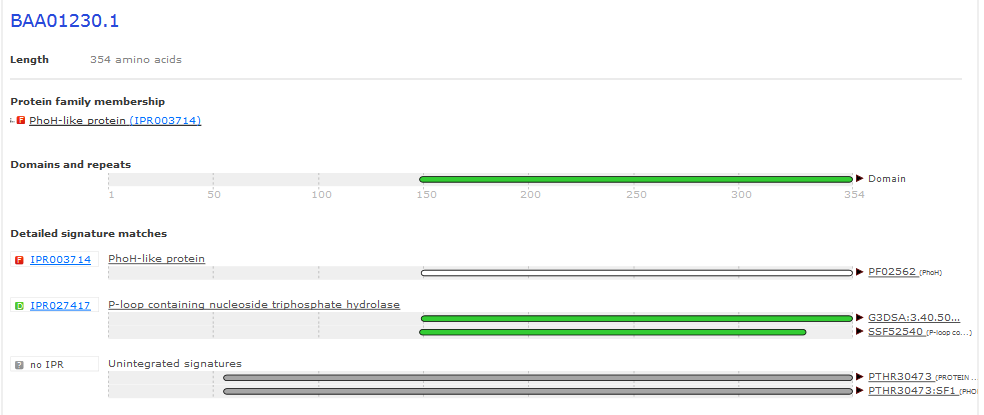
Note that the region that was different in the alignment is also missing the PIN signature.
What is gp14?¶
- We still don’t know!
- It’s probably not just like PhoH, because:
- The PhoH conserved domain is really a predicted ATPase. There are all kinds of ATPases, many proteins can use ATP to drive reactions.
- The “real” PhoH does not contain a PIN domain
So what would I annotate gp14 as?¶
- Bacterial homologs are mostly annotated as ATPase, which is more
conservative
- Probably more accurate but less specific
- Could also annotate as putative PIN ribonuclease domain protein
- None of this is very standardized, and there is sometimes no right answer
- This is why it is important to put the rationale for your annotation in the comments field!



