Class 4 Slides: Signals, Ends and Lysis Cassettes¶
Signals¶
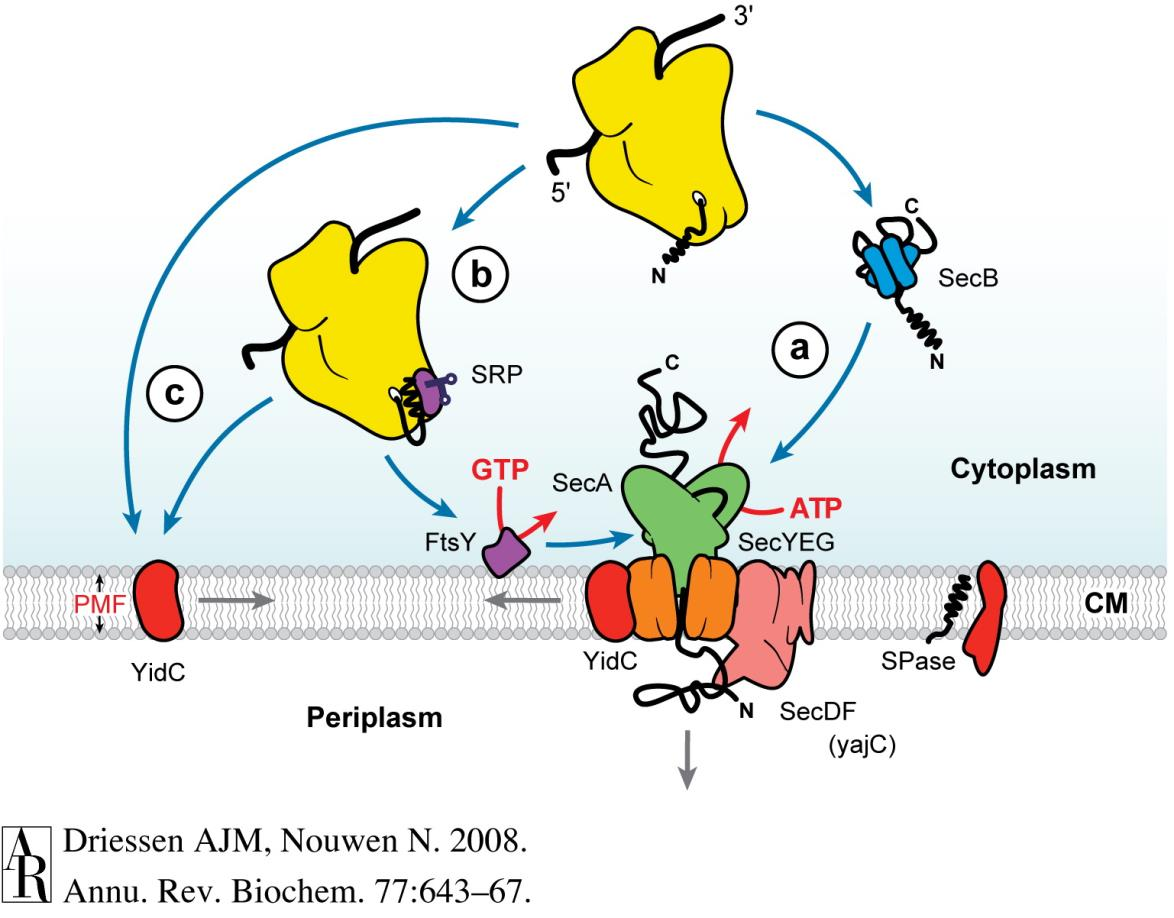
The sec system for protein secretion¶
- All integral membrane proteins, periplasmic proteins and OM proteins must be secreted through the IM
- The major mechanism for this is the sec system
- The sec system transports unfolded proteins through the IM
The sec system for protein secretion¶
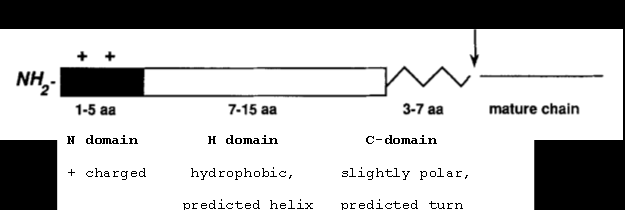
Signal Sequences: SPI¶
- Control secretion and localization of a protein
- Secretion signal sequences are always located at the N-terminus of a protein
- The most common sec signal sequence type
Lipoproteins¶
Proteins with a slightly different signal sequence are recognized by SPII instead of SPI, and are lipid modified with a diacylglycerol on the S atom and a third FA on the N-terminal amine, to make a lipoprotein
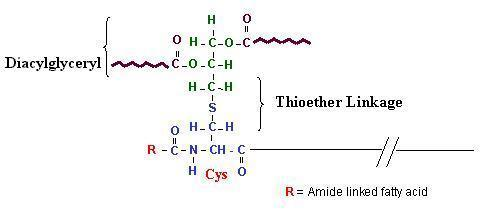
Signal Sequences: SPII¶
- Like the SPI signal but contains a lipobox motif at the end of the C domain
- The lipobox is followed by a cysteine, which is lipid modified
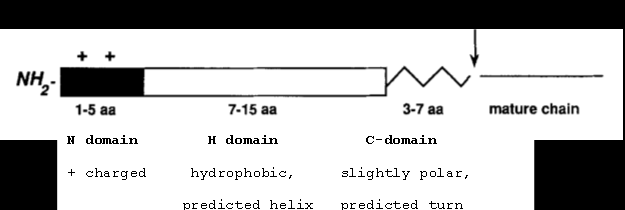
The site marked with an arrow between “3-7 aa” and “Mature chain” is the cleavage site.
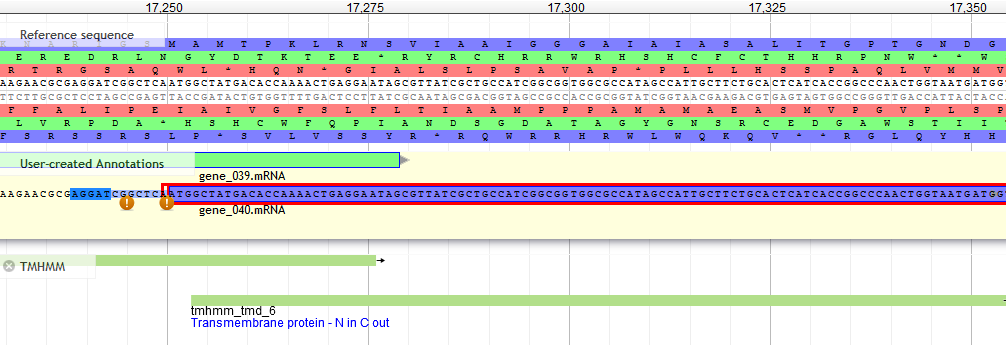
Transmembrane Domains¶
- In general are 15 to 22 AA
- Hydrophobic, uncharged (ile, leu, val, phe, tyr, trp, met)
- Alpha-helical

Example of a TMD. Note that the TMD regions are generally uncharged, as they exist inside the lipid bilayer in the cell. In order to stay integrated into the membrane, they must be chemically similar to the surrounding lipids
TMD Snorkeling¶
- Lysine residues can “snorkel” to the bilayer surface three residues or less from the end of a TMD helix
- Adjacent positive and negative charges can neutralize each other in the same helix
- These features can confuse TMD prediction
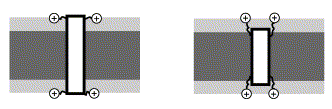
“Positive Inside” Rule¶
- The inner membrane carries a net positive charge on the outside surface
- Membrane proteins will take a topology that keeps net negative charges on the outside, and net positive charges on the inside
- Note that TMD prediction tools often mistake SPI and SPII signals for N-terminal TMDs!

Signal Anchor Release (SAR) Domains¶
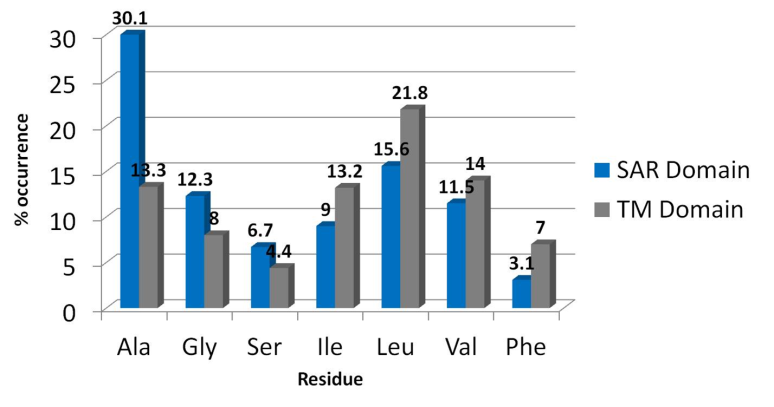
- These are like normal TMD’s but are enriched in weakly hydrophobic residues like gly, ala, ser
- Inserted into the membrane like normal TMD but will release from the membrane at a low rate
- These can also be missed by TMD prediction software
Lysis Cassettes¶
- All Caudovirales phages have lysis genes
- These genes are often arranged in a lysis cassette, containing the holin
(and antiholin), endolysin and spanin complex
- In some phages (most notably T4 and its relatives), the lysis genes are spread over the genome which makes annotating them more difficult
- When annotating, start with the assumption that you have a lysis cassette
Lysis Cassette Annotation: Endolysin¶
- Step 1: find your endolysin Endolysins of all known classes are
well-conserved and should be easy to find
- Canonical phage DB
- InterProSan
- UniRef
- Holins and spanins are generally not conserved and you are unlikely to find them directly by BLAST or InterProScan
Lysis Cassette Annotation: Critical Evaluation¶
- Step 2: Look at your endolysin
- If your endolysin has a single N-terminal TMD, it is a putative SAR endolysin
- If not, it is a putative soluble endolysin
- Examine the InterPro and PhageDB results and try to classify your endolysin by its catalytic type
Lysis Cassette Annotation: Critical Evaluation¶
- A SAR endolysin’s TMD is weakly hydrophobic, enriched in ala, gly and ser residues
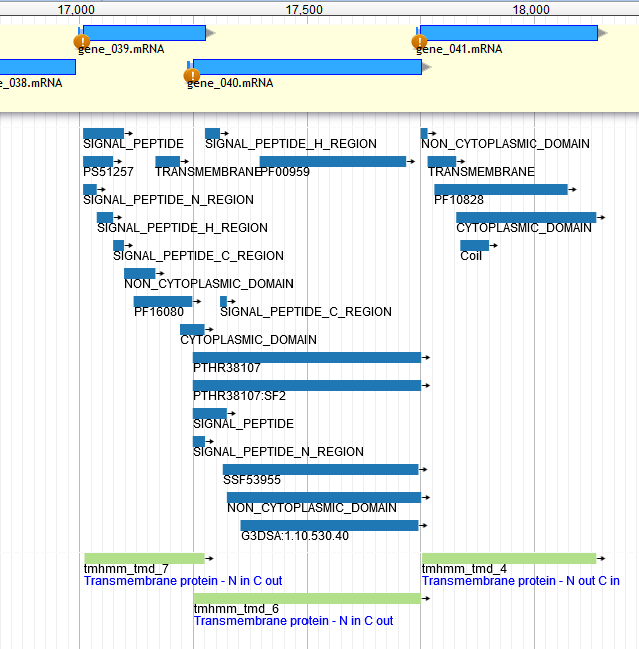
Lysis Cassette Annotation: Rounding Up the Usual Suspects¶
- Step 3: look up and downstream for TMD-containing proteins
- InterProScan
- TMHMM
- One of these is probably the holin, one is probably the spanin
- You may have several TMD-containing proteins nearby
- Holin and antiholin as separate genes
- IM spanin and OM spanin as separate genes
- Other unrelated proteins (red herrings)
- You may have no TMD-containing proteins nearby
- Phage has a distributed lysis cassette
- The endolysin itself may contain a TMD
- SAR endolysin
Lysis Cassette Annotation: Identifying the Holin¶
- Step 4: Look for your holin
- A “classic” holin is a small protein containing 2-3 TMDs
- Unless it is a Class III holin (as in T4), which has only 1 N- terminal TMD
- Multiple adjacent holin-like proteins may indicate a separated
holin-antiholin system
- Cannot distinguish between holin and antiholin based on sequence (annotate as “putative holin or antiholin”)
Lysis Cassette Annotation: IM Spanin¶
- Step 5: Look for your IM spanin
- The IM spanin has a single N-terminal TMD
- The identity of the IM spanin is confirmed by the presence of an OM
spanin gene embedded inside or directly downstream
- Embedded OM spanins were probably not annotated
- Workflow for OM spanin finding coming soon
Ends¶
A word on phage DNA packaging¶
Five major types of phage packaging:
- Cos (3’ or 5’ overhang)
- Pac (with or without a defined start site)
- Terminal repeats (short or long)
- Host DNA (rare, only found in Mu-like phages)
- Terminal proteins (rare, only found in phi29-like phages)
Types of Phage DNA Termini¶
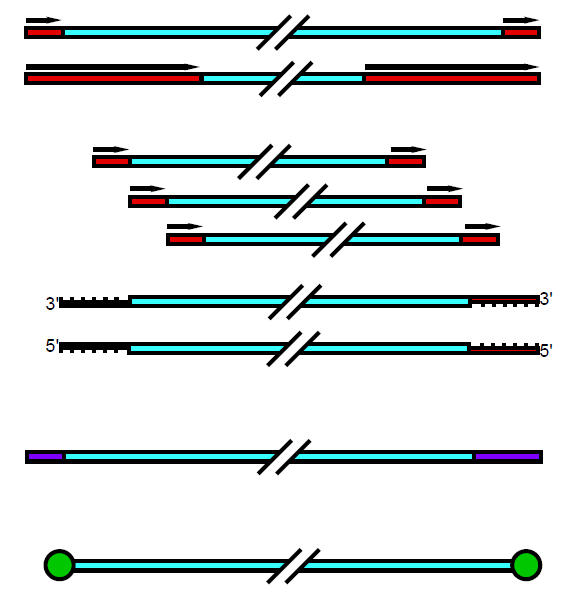
Five main types:
- Terminal Repeats
- Long (coding)
- Short (non-coding)
- Pac (headfull)
- Defined or undefined packaging initiation
- Cos (overhangs)
- 5’ or 3’
- Host DNA (Mu)
- Terminal protein (phi29)
DNA Packaging as a Two-Step Process¶
Packaging Initiation
- Site-specific
- Cos
- Pac
- Terminal Repeat
- Mu-like
- Non-site-specific
- Pac
**Packaging Termination
- Site Specific
- Cos
- Terminal Repeat
- Non-site-specific
- Pac
- Mu-like
- Some TR?
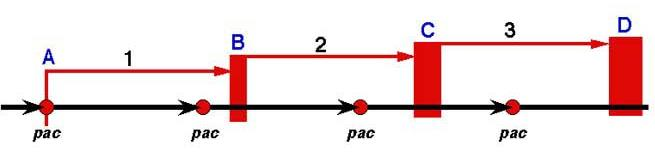
Pac-type Packaging¶
- Headfull packaging can include two similar packaging styles
- Initiation can be specific OR non-specific (or something in between) * Termination is always headfull (not site-specific)
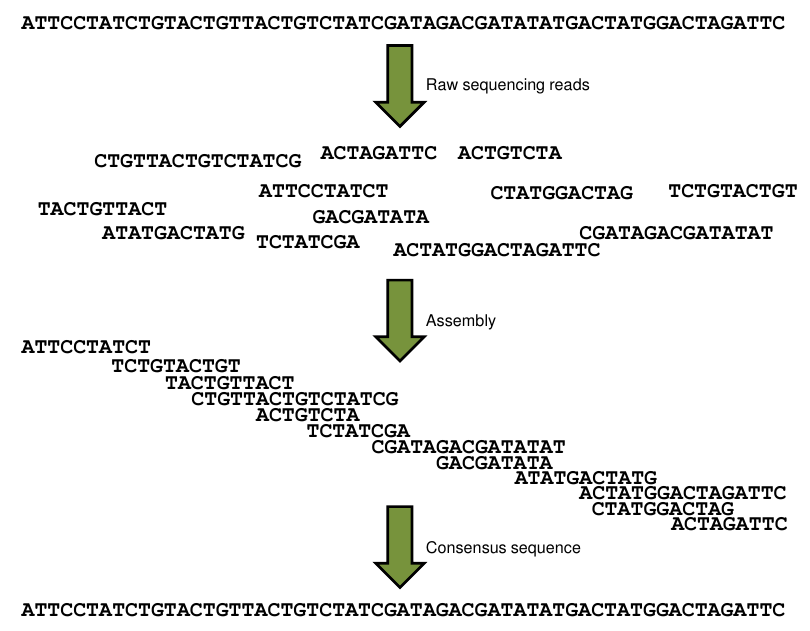
DNA Sequence Assembly¶
Types of Phage DNA Termini¶
| Type | Description | Examples |
|---|---|---|
| Short (hundreds of bp) or long (several kb) direct terminal repeats | Genomes with this architecture typically result in circular assemblies following sequencing, as identical sequence reads from the termini collapse into a single region in the assembled contig | T7 (short), SPO1 (long) |
| Permuted | These genomes also yield circular assemblies, depending on the degree of permutation. In significantly permuted genomes, there is no “true” terminus, and the genome is opened at an arbitrary location, typically at the terminase genes. | T4 |
| Non-permuted with cohesive ends | Virions contain nonpermuted genomes with short (<20 bp) 5’ or 3’ single-stranded DNA overhang. The termini of the assembled contig typically represent the genome termini, however the extent of the overhangs must be determined experimentally. | lambda |
| Random host DNA at termini | When sequenced by traditional dideoxy sequencing at relatively low (approximately 8-fold) coverages, reads containing host DNA sequence could be observed in the assemblies. In next-generation sequencing, such mismatching reads are often automatically discarded during assembly and may be missed without careful examination of sequencing data. | Mu |
| Covalently attached protein | Depending on how the DNA was prepared, terminal fragments with attached protein may be lost to sequencing, resulting in an incomplete genome. | phi29 |
Large Terminase¶
- The large terminase (TerL) determines packaging type
- TerL proteins with the same packaging type tend to be more related to each other